As an API
Pandora2D API usage
Pandora2D provides a full python API which can be used to compute disparity maps as shown in this basic example:
import numpy as np
import pandora
import pandora2d
from pandora2d.state_machine import Pandora2DMachine
from pandora2d import check_configuration, common
from pandora2d.img_tools import create_datasets_from_inputs
# path to save disparity maps
path_output = "./res"
# Paths to left and right images
img_left_path = "data/left.tif"
img_right_path = "data/right.tif"
# image configuration
image_cfg = {
'input': {
'left': {
'img': img_left_path,
'nodata': np.nan,
},
'right': {
'img': img_right_path,
'nodata': np.nan,
},
"col_disparity": [-2, 2],
"row_disparity": [-2, 2],
}
}
# read images
image_datasets = create_datasets_from_inputs(input_config=image_cfg["input"])
# instantiate Pandora2D Machine
pandora2d_machine = Pandora2DMachine()
# define pipeline configuration
user_pipeline_cfg = {
'pipeline':{
"matching_cost" : {
"matching_cost_method": "zncc",
"window_size": 5,
},
"disparity": {
"disparity_method": "wta",
"invalid_disparity": -9999
},
"refinement" : {
"refinement_method" : "interpolation"
}
}
}
# check the configurations and sequences steps
pipeline_cfg = check_configuration.check_pipeline_section(user_pipeline_cfg, pandora2d_machine)
# prepare the machine
pandora2d_machine.run_prepare(
image_datasets.left,
image_datasets.right
)
# trigger all the steps of the machine at ones
dataset = pandora2d.run(
pandora2d_machine,
image_datasets.left,
image_datasets.right,
pipeline_cfg
)
# save dataset
common.save_dataset(dataset, path_output)
Pandora2D’s data
Images
Pandora2D reads the input images before the stereo computation and creates two datasets, one for the left and one for the right image which contain the data’s image, data’s mask and additionnal information.
Example of an image dataset
Dimensions: (col: 450, row: 375)
Coordinates:
* col (col) int64 0 1 2 3 4 5 6 7 8 ... 442 443 444 445 446 447 448 449
* row (row) int64 0 1 2 3 4 5 6 7 8 ... 367 368 369 370 371 372 373 374
* band_disp (band_disp) <U3 'min' 'max'
Data variables:
im (row, col) float32 88.0 85.0 84.0 83.0 ... 176.0 180.0 165.0 172.0
msk (row, col) int16 0 0 0 0 0 0 0 0 0 0 0 0 ... 0 0 0 0 0 0 0 0 0 0 0
col_disparity (band_disp, row, col) int64 -2 -2 -2 -2 ... 2 2 2 2
row_disparity (band_disp, row, col) int64 -2 -2 -2 -2 ... 2 2 2 2
Attributes:
no_data_img: 0
crs: None
transform: | 1.00, 0.00, 0.00|| 0.00, 1.00, 0.00|| 0.00, 0.00, 1.00|
valid_pixels: 0
no_data_mask: 1
col_disparity_source: [-2, 2]
row_disparity_source: [-2, 2]
Two data variables are created in this dataset:
* *im*: contains input image data
* *msk*: contains input mask data + no_data of input image
Note
This example comes from a dataset created by Pandora’s reading function. Dataset attributes valid_pixels and no_data_mask cannot be modified with this function, as they are defined by the msk data convention. For an API user who wants to create its own dataset without using Pandora’s reading function, it is possible to declare its own mask convention with these attributes:
no_data_img : value of no_data in input image
valid_pixels: value of valid pixels in input mask
no_data_mask: value of no_data pixel in input mask
Cost volumes
Pandora2D will then store all the cost volumes together in a 4D (dims: row, col, disp_col, disp_row) xarray.DataArray named cost_volumes. When matching is impossible, the matching cost is set to np.nan.
<xarray.Dataset>
Dimensions: (col: 3, disp_col: 2, disp_row: 2, row: 3)
Coordinates:
row (row) int64 0 1 2
col (col) int64 0 1 2
disp_col (disp_col) int64 -1 0
disp_row (disp_row) int64 -1 0
Data variables:
cost_volumes (row, col, disp_col, disp_row) float32 nan nan ... 4.0
Attributes:
measure: sad
subpixel: 1
offset_row_col: 0
window_size: 1
type_measure: min
cmax: 10004
crs: None
transform: | 1.00, 0.00, 0.00|| 0.00, 1.00, 0.00|| 0.00, 0.00, ...
Disparity map
The Disparity computation step generates two disparity maps in cost volume geometry. One named row_map for the vertical disparity and one named col_map for the horizontal disparity. These maps are float32 type 2D xarray.DataArray, stored in a xarray.Dataset.
<xarray.Dataset>
Dimensions: (col: 450, row: 375)
Coordinates:
* row (row) int64 0 1 2 3 4 5 6 7 8 ... 367 368 369 370 371 372 373 374
* col (col) int64 0 1 2 3 4 5 6 7 8 ... 442 443 444 445 446 447 448 449
Data variables:
row_map (row, col) float32 nan nan nan nan nan nan ... nan nan nan nan nan
col_map (row, col) float32 nan nan nan nan nan nan ... nan nan nan nan nan
Border management
Left image
Pixels of the left image for which the measurement thumbnail protrudes from the left image are set to 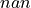 on the cost volume.
For a similarity measurement with a 5x5 window, these incalculable pixels in the left image correspond
to a 2-pixel crown at the top, bottom, right and left, and are represented by the offset_row_col attribute in
the xarray.Dataset.
on the cost volume.
For a similarity measurement with a 5x5 window, these incalculable pixels in the left image correspond
to a 2-pixel crown at the top, bottom, right and left, and are represented by the offset_row_col attribute in
the xarray.Dataset.
Right image
Because of the disparity range choice, it is possible that there is no available point to scan on the right image.
In this case, matching cost cannot be computed for this pixel and the value will be set to 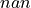 .
Then bit 1 will be set : The point is invalid: the disparity interval to explore is
absent in the right image and the point disparity will be set to invalid_disparity.
Moreover, everytime Pandora2D shifts the right image it introduces a new line set at nodata_right value. The matching
cost cannot be computed for this line to.
.
Then bit 1 will be set : The point is invalid: the disparity interval to explore is
absent in the right image and the point disparity will be set to invalid_disparity.
Moreover, everytime Pandora2D shifts the right image it introduces a new line set at nodata_right value. The matching
cost cannot be computed for this line to.